
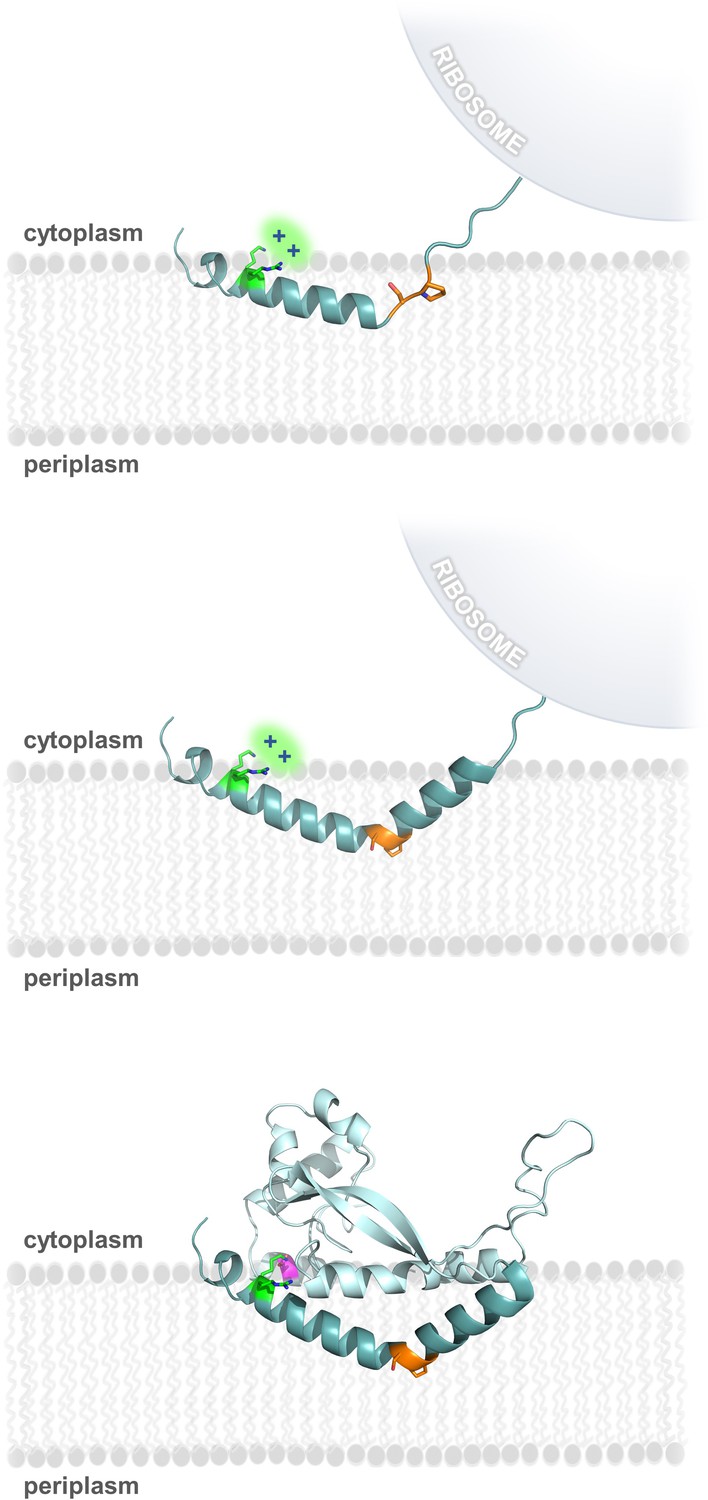
Such residues are called hydrophobic or "water-fearing." As a result, the interaction between the water-loving and water-fearing residues of the protein and the surrounding watery fluids helps drive protein folding and stabilizes the protein's final structure. Non-polar residues, on the other hand, tend to be found packed within the protein core away from the polar aqueous fluid. As a result, polar residues on proteins are called hydrophilic, or "water-loving." In aqueous fluids, amino acid residues that have polar sidechains - components that can have a charge under certain physiological conditions or that participate in hydrogen bonding - tend to be located on the surface of the protein where they can interact with water, which has negatively and positively side charges to its molecule. The resulting models have been shown to accurately represent the structure the sequence will likely assume in nature.ĭetermining the structure of transmembrane proteins is difficult because portions of transmembrane proteins must pass though the membrane's interior, which is made of oily fats called lipids. It is not unusual for the program to create tens of thousands of model structures for an amino acid sequence and then identify the ones with lowest energy state. The Rosetta program used by Lu and his colleagues can predict the structure of a protein by taking into account these interactions and calculating the lowest overall energy state. Ultimately, the protein assumes the shape that best balances out all these factors so that the protein achieves the lowest possible energy state. The architecture of a protein is crucial because a protein's structure determines its function.Ī protein's shape forms from complex interactions between the amino acids that make up the protein chain and between the amino acids and the surrounding environment. In the new study, Lu and his coworkers used a computer program, developed in the Baker lab and called Rosetta, that can predict the structure a protein will fold into after it has been synthesized. Because they act while embedded within the cellular membrane, transmembrane proteins have proven to be more difficult to study than proteins that operate in the watery solution that make up the cells' cytoplasm or in the extracellular fluid. Peilong Lu, a senior fellow in the Baker lab, is the paper's lead author.īut understanding how transmembrane proteins are put together and how they work has proved challenging. The research is reported in the March 1 issue of the journal Science. "Our results pave the way for the design of multispan membrane proteins that could mimic proteins found in nature or have entirely novel structure, function and uses," said David Baker, a University of Washington School of Medicine professor biochemistry and director of the UW Institute of Protein Design who led the project. Now researchers are looking at designing the transmembrane proteins themselves to perform specific tasks. Because of such roles, many drugs are designed to target transmembrane proteins and alter their function.
Some transmembrane proteins receive or transmit cell signals. For example, many naturally occurring transmembrane proteins act as gateways for the movement of specific substances across a biological membrane. They are essential for them to function normally. In the living world, transmembrane proteins are found embedded in the membrane of all cells and cellular organelles. Included in the publication is the profiling of a range of small molecule compounds and biological stimuli which demonstrated the potential of the method to identify and study membrane – bound protein targets. , CETSA ® Explore was combined with selective enrichment of glycosylated cell surface proteins.

This is hugely valuable to drug discovery researchers in that by demonstrating how CETSA ® can be used for membrane-bound proteins, such as ion channels and transporters, scientists now have an alternative to biochemical or biophysical methods. This stud y describe s the thorough experimental work undertaken to develop assay protocols for the more challenging multi-pass transmembrane proteins.
Multipass transmembrane protein how to#
Ĭase studies on how to deploy CETSA ® on insoluble protein targets such as membrane proteins have been published, for example by Kaw atkar et al. Nevertheless, CETSA ® has successfully been applied to a number of membrane- associated proteins including GPCR s and transporter s. CETSA ® has primarily been applied to soluble proteins which, upon heating, denaturate to form insoluble aggregates by exposing hydrophobic surfaces.


 0 kommentar(er)
0 kommentar(er)
